Shortest Paths
Weighted Graphs
- In a weighted graph, each edge has an associated numerical value, called the weight of the edge
- Edge weights may represent, distances, costs, etc.
- Example:
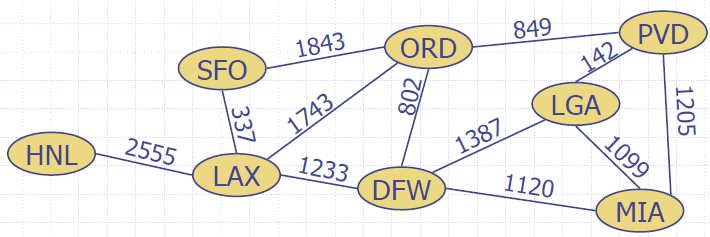
- In a flight route graph, the weight of an edge represents the distance in miles between the endpoint airports
Shortest Path Problem
- Given a weighted graph and two vertices u and v, we want to find a path of minimum total weight between u and v.
- Length of a path is the sum of the weights of its edges.
- Example:
- Shortest path between Providence and Honolulu
- Applications: Internet packet routing, Flight reservations, Driving directions
Shortest Path Properties
- Property 1:
- A subpath of a shortest path is itself a shortest path
- Property 2:
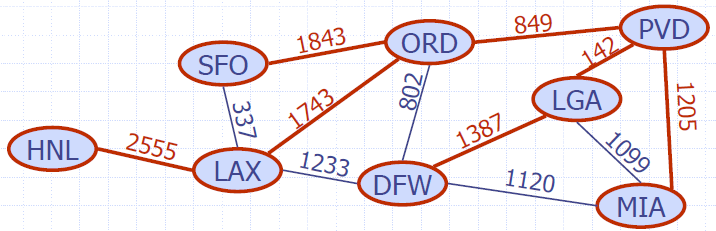
- There is a tree of shortest paths from a start vertex to all the other vertices
- Example:
- Tree of shortest paths from Providence
Dijkstra's Algorithm
- The distance of a vertex v from a vertex s is the length of a shortest path between s and v
- Dijkstra’s algorithm computes the distances of all the vertices from a given start vertex s
- Assumptions:
- the graph is connected
- the edges are undirected
- the edge weights are nonnegative
- We grow a “cloud” of vertices, beginning with s and eventually covering all the vertices
- We store with each vertex v a label d(v) representing the distance of v from s in the subgraph consisting of the cloud and its adjacent vertices
- At each step
- We add to the cloud the vertex u outside the cloud with the smallest distance label, d(u)
- We update the labels of the vertices adjacent to u
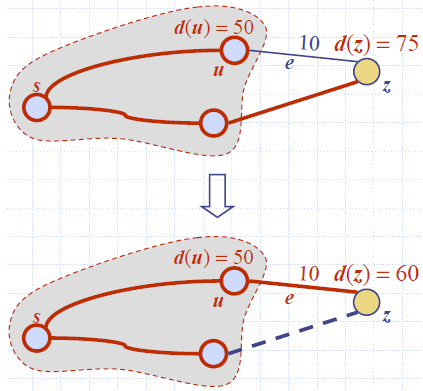
Edge Relaxation
- Consider an edge e = (u,z) such that
- u is the vertex most recently added to the cloud
- z is not in the cloud
- The relaxation of edge e updates distance d(z) as follows:
- d(z) ← min{d(z),d(u) + weight(e)}
Example
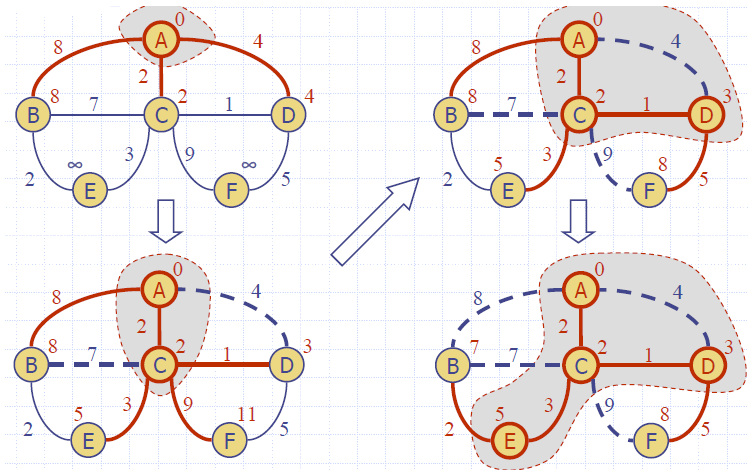
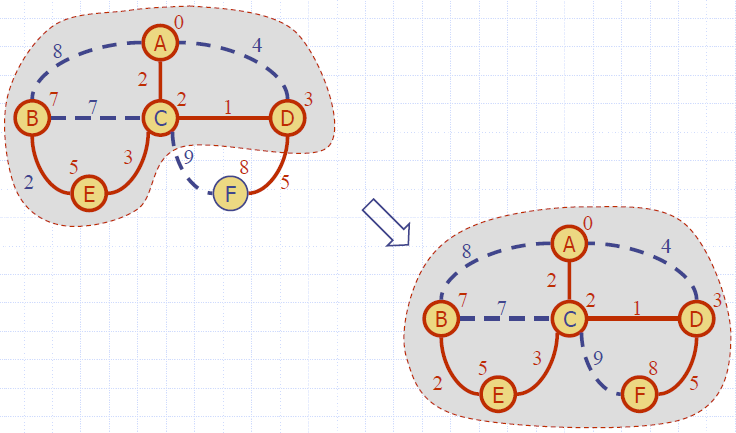
Dijkstra’s Algorithm
- A priority queue stores the vertices outside the cloud
- Key: distance
- Element: vertex
- Locator-based methods
- insert(k,e) returns a locator
- replaceKey(l,k) changes the key of an item
- We store two labels with each vertex:
- Distance (d(v) label)
- locator in priority queue
Algorithm DijkstraDistances(G, s)
Q ← new heap-based priority queue
for all v ∈ G.vertices()
if v = s
setDistance(v, 0)
else
setDistance(v, ∞)
l ← Q.insert(getDistance(v), v)
setLocator(v,l)
while ¬Q.isEmpty()
u ← Q.removeMin()
for all e ∈ G.incidentEdges(u)
{ relax edge e }
z ← G.opposite(u,e)
r ← getDistance(u) + weight(e)
if r < getDistance(z)
setDistance(z,r)
Q.replaceKey(getLocator(z),r)
Analysis
- Graph operations
- Method incidentEdges is called once for each vertex
- Label operations
- We set/get the distance and locator labels of vertex z O(deg(z)) times
- Setting/getting a label takes O(1) time
- Priority queue operations
- Each vertex is inserted once into and removed once from the priority queue, where each insertion or removal takes O(log n) time
- The key of a vertex in the priority queue is modified at most deg(w) times, where each key change takes O(log n) time
- Dijkstra’s algorithm runs in O((n + m) log n) time provided the graph is represented by the adjacency list structure
- Recall that Σv deg(v) = 2m
- The running time can also be expressed as O(m log n) since the graph is connected
Code
package examples;
import graphLib.*;
import graphTool.Algorithm;
import graphTool.Attribute;
import graphTool.GraphTool;
import java.awt.Color;
import java.io.Serializable;
import java.util.ArrayList;
import java.util.Arrays;
import java.util.Iterator;
import java.util.LinkedList;
import java.util.Random;
public class GraphExamples<V,E> {
@Algorithm(vertex=true)
public void dijkstra(Graph<V,E> g,Vertex<V> s, GraphTool<V,E> t){
t.show(g);
MyPriorityQueue<Double, Vertex<V>> pq = new MyPriorityQueue<>();
Iterator<Vertex<V>> it = g.vertices();
while(it.hasNext()){
Vertex<V> v = it.next();
v.set(Attribute.DISTANCE,Double.POSITIVE_INFINITY);
v.set(Attribute.string,"inf");
Locator<Double,Vertex<V>> loc = pq.insert(Double.POSITIVE_INFINITY,v);
v.set(Attribute.PQLOCATOR,loc);
t.show(g);
}
s.set(Attribute.string,"0");
s.set(Attribute.DISTANCE,0.0);
t.show(g);
pq.replaceKey((Locator<Double,Vertex<V>>)s.get(Attribute.PQLOCATOR),0.0);
while( ! pq.isEmpty()){
Vertex<V> u = pq.removeMin().element();
u.set(Attribute.color,Color.GREEN);
if (u.has(Attribute.DISCOVERY)){
Edge<E> e = (Edge<E>)u.get(Attribute.DISCOVERY);
e.set(Attribute.color,Color.GREEN);
t.show(g);
}
Iterator<Edge<E>> eIt;
if (g.isDirected()) eIt = g.incidentInEdges(u);
else eIt = g.incidentEdges(u);
while (eIt.hasNext()){
Edge<E> e = eIt.next();
double weight = 1.0;
if (e.has(Attribute.weight)) {
weight = (Double)e.get(Attribute.weight);
}
Vertex<V> z = g.opposite(e, u);
double newDist = (Double) u.get(Attribute.DISTANCE);
if (newDist < (Double)z.get(Attribute.DISTANCE)){
z.set(Attribute.DISTANCE,(Double)newDist);
z.set(Attribute.string,""+newDist);
z.set(Attribute.DISCOVERY,e);
z.set(s,u);
pq.replaceKey((Locator<Double,Vertex<V>>)z.get(Attribute.PQLOCATOR),newDist);
t.show(g);
}
}
}
t.show(g);
}
public static void main(String[] args) {
IncidenceListGraph<String,String> g =
new IncidenceListGraph<>(true);
GraphExamples<String,String> ge = new GraphExamples<>();
Vertex vA = g.insertVertex("A");
vA.set(Attribute.name,"A");
Vertex vB = g.insertVertex("B");
vB.set(Attribute.name,"B");
Vertex vC = g.insertVertex("C");
vC.set(Attribute.name,"C");
Vertex vD = g.insertVertex("D");
vD.set(Attribute.name,"D");
Vertex vE = g.insertVertex("E");
vE.set(Attribute.name,"E");
Vertex vF = g.insertVertex("F");
vF.set(Attribute.name,"F");
Vertex vG = g.insertVertex("G");
vG.set(Attribute.name,"G");
Edge e_a = g.insertEdge(vA,vB,"AB");
Edge e_b = g.insertEdge(vD,vC,"DC");
Edge e_c = g.insertEdge(vD,vB,"DB");
Edge e_d = g.insertEdge(vC,vB,"CB");
Edge e_e = g.insertEdge(vC,vE,"CE");
e_e.set(Attribute.weight,2.0);
Edge e_f = g.insertEdge(vB,vE,"BE");
e_f.set(Attribute.weight, 7.0);
Edge e_g = g.insertEdge(vD,vE,"DE");
Edge e_h = g.insertEdge(vE,vG,"EG");
e_h.set(Attribute.weight,3.0);
Edge e_i = g.insertEdge(vG,vF,"GF");
Edge e_j = g.insertEdge(vF,vE,"FE");
GraphTool t = new GraphTool(g,ge);
}
}
Extension of Dijkstra
- Using the template method pattern, we can extend Dijkstra’s algorithm to return a tree of shortest paths from the start vertex to all other vertices
- We store with each vertex a third label:
- parent edge in the shortest path tree
- In the edge relaxation step, we update the parent label
Algorithm DijkstraShortestPathsTree(G, s)
…
for all v ∈ G.vertices()
…
setParent(v, ∅)
…
for all e ∈ G.incidentEdges(u)
{ relax edge e }
z ← G.opposite(u,e)
r ← getDistance(u) + weight(e)
if r < getDistance(z)
setDistance(z,r)
setParent(z,e)
Q.replaceKey(getLocator(z),r)
Why Dijkstra’s Algorithm Works
- Dijkstra’s algorithm is based on the greedy method. It adds vertices by increasing distance.
- Suppose it didn’t find all shortest distances. Let F be the first wrong vertex the algorithm processed.
- When the previous node, D, on the true shortest path was considered, its distance was correct.
- But the edge (D,F) was relaxed at that time!
- Thus, so long as d(F)>d(D), F’s distance cannot be wrong. That is, there is no wrong vertex.
Why It Doesn’t Work for Negative-Weight Edges
- Dijkstra’s algorithm is based on the greedy method. It adds vertices by increasing distance.
- If a node with a negative incident edge were to be added late to the cloud, it could mess up distances for vertices already in the cloud.
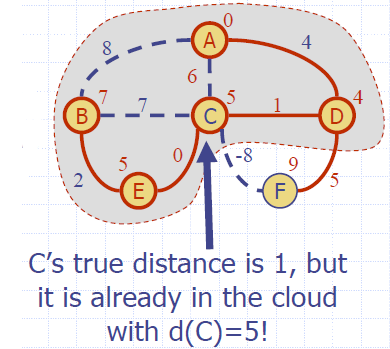
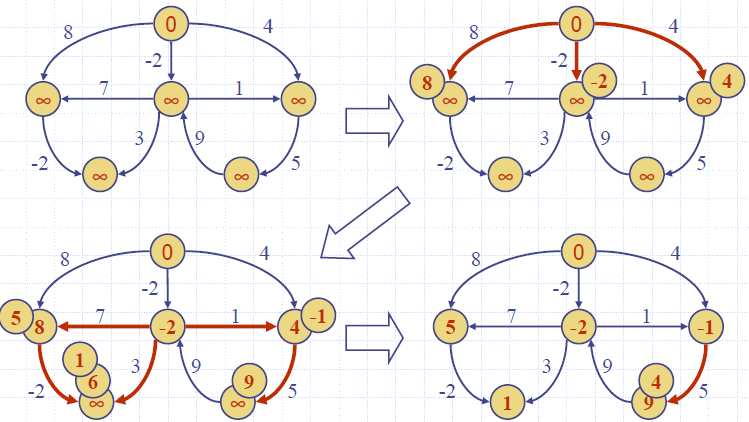
Bellman-Ford Algorithm
- Works even with negativeweight edges
- Must assume directed edges (for otherwise we would have negativeweight cycles)
- Iteration i finds all shortest paths that use i edges.
- Running time: O(nm).
- Can be extended to detect a negative-weight cycle if it exists
Algorithm BellmanFord(G, s)
for all v ∈ G.vertices()
if v = s
setDistance(v, 0)
else
setDistance(v, ∞)
for i ← 1 to n-1 do
for each e ∈ G.edges()
{ relax edge e }
u ← G.origin(e)
z ← G.opposite(u,e)
r ← getDistance(u) + weight(e)
if r < getDistance(z)
setDistance(z,r)
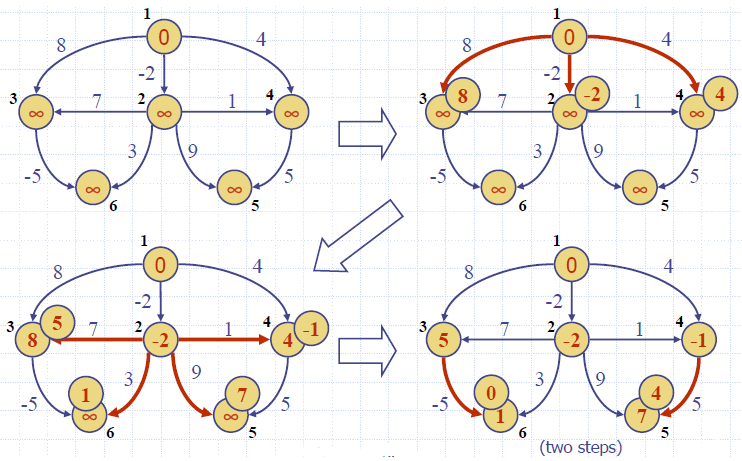
Example
DAG-based Algorithm
- Works even with negative-weight edges
- Uses topological order Doesn’t use any fancy data structures
- Is much faster than Dijkstra’s algorithm
- Running time: O(n+m).
Algorithm DagDistances(G, s)
for all v ∈ G.vertices()
if v = s
setDistance(v, 0)
else
setDistance(v, ∞)
Perform a topological sort of the vertices
for u ← 1 to n do {in topological order}
for each e ∈ G.outEdges(u)
{ relax edge e }
z ← G.opposite(u,e)
r ← getDistance(u) + weight(e)
if r < getDistance(z)
setDistance(z,r)
Example
All-Pairs Shortest Paths
- Find the distance between every pair of vertices in a weighted directed graph G.
- We can make n calls to Dijkstra’s algorithm (if no negative edges), which takes O(nmlog n) time.
- Likewise, n calls to Bellman-Ford would take O(n2m) time.
- We can achieve O(n3) time using dynamic programming (similar to the Floyd-Warshall algorithm).
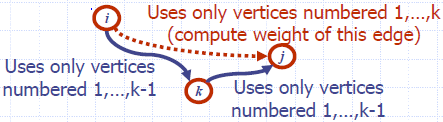
Algorithm AllPair(G) {assumes vertices 1,…,n}
for all vertex pairs (i,j)
if i = j
D0[i,i] ← 0
else if (i,j) is an edge in G
D0[i,j] ← weight of edge (i,j)
else
D0[i,j] ← + ∞
for k ← 1 to n do
for i ← 1 to n do
for j ← 1 to n do
Dk[i,j] ← min{Dk-1[i,j], Dk-1[i,k]+Dk-1[k,j]}
return Dn